|
Beckman Research Institute of the City of Hope, Duarte, CA 91010, USA. Introduction Ever since the X-ray crystallographic analysis of
a class I major histocompatibility complex (MHC) antigen revealed
the presence of an alien peptide fragment sandwiched between its
two parallel IXhelices [1], the immunological self became a multitude
of such peptide fragments, usually 15-20 residues long, derived
from host proteins after intracellular processing. For the mainly
intra thymic education of self to cytotoxic T cells, these fragments
are presented in association with class I MHC antigens, while for
the education of helper T cells, they are presented with class II
MHC antigens. For those who believe that proteins represent random
assemblages of 20 amino acid residues, the above manner of presentation
of self poses no problem, for 15- 20 residues long peptide fragments
represent an astronomical variety of 2015-202°. With this much variety,
homologous peptide fragments are to be found only among proteins
related by the propinq uity of their descents. Thus, viral and other
pathogenic peptide fragments would be distinct from most of the
host peptide fragments. The purpose of this paper is to show that
the above is far from the truth. Many peptide fragments are syntactical
in construction, and are therefore to be found in many totally unrelated
proteins. The average amino acid composition deduced from 18383
entries in Database is as follows: (1) The top four residues, Leu,
Ala, Gly, and Ser, in this order, comprise 32% of the total, and
(2) the bottom four residues, His, Met, Cys, and Trp, in this order,
comprise only 7% of the total. All 20 homodipeptides occurred at
above their expected rates, thus, homodipeptides in the average
protein acounted for 14% of its length. While the LeuLeu homodipeptide
was the most numerous of the 400 dipeptides, the second in rank
was Leu- Val, occurring at nearly twice the expected rate, while
its reciprocal Val-Leu was only one-third as numerous [2]. The above
can be viewed as a rudimentary indication of syntatic structures
in amino acid sequences. In order to expand on this theme, I have
chosen four totally unrelated proteins as representatives of the
warm-blooded verterate host. They are: (1) human ET .REC (estrogen
receptor), 595 residues long [3]; (2) chicken C-SRC (tyrosine kinase),
533 residue long [4]; (3) human S.ALB (serum albumin), 585 residue
long [5]; and (4) human PGK (phospholglycerate kinase) 415 residue
long [6]. Lys-Leu- and Leu-Lys-Containing Oligopeptides in Four
Host Proteins We shall now start our inquiry by choosing a pair
of Leciprocal dipeptides, LysLeu and Leu-Lys. According to the aforementioned
extensive survey of 18383 entries in Database, Lys-Leu occurred
at about the expected rate, while the incidence of its reciprocal
Leu-Lys was slightly less [2]. In the case of four host proteins
totalling 2128 residues, there were only 12 Lys-Leu and 13 Leu Lys.
Yet, three of the 12 Lys-Leu dipeptides appeared as Val-Lys-Leu
and two of them as Ser-Lys-Leu tripeptides. These are indisputable
cases of preferential associations, for the most abundant tripeptide
ending in Lys-Leu should have been the palindromic Leu-Lys-Leu which,
on a random basis, had the expected incidence of 1.08. The fact
is that there was not a single Leu-Lys-Leu tripeptide among the
four proteins. As to its carboxyl end partners, the Lys-Leu dipeptide
showed a distinct preference for Val and the next Gly, for there
were four Lys-Leu- Val and two Lys-Leu-Gly. Accordingly, it was
no surprise that two totally unrelated proteins, C-SRC and S.ALB,
shared a pair of homologous tetrapeptides. Lys-Leu- ValGIn and Lys-Leu-Val-Asn,
as shown in Fig. 1 a. As to the 13 Leu-Lys found in four host proteins,
this dipeptide showed a definite preference to associate with Phe
as its amino terminal partner (four PheLeu-Lys in C-SCR, S.ALB,
and PGK) and a preference for Ser as its carboxyl terminal partner
(three Leu-Lys-Ser in ET.REC and PGK). Accordingly, a pair of homologous
pentapeptides containing Leu-Lys was shared between ET.REC and PGK
and a pair of identical tetrapeptides, Thr-Phe-Leu-Lys, between
S.ALB and PGK. As to two pairs of homologous tetrapeptides containing
Leu-Lys or IleLys, the first was shared by S.ALB and PGK and the
second by ET.REC and CSRC, as also shown in Fig. 1 a. Lys-Leu- and Leu-Lys-Containing Oligopeptides in Two Influenza A Virus Hemagglutinins As it has now become clear that totally unrelated host proteins
commonly share homologous and identical penta- and tetrapeptides
between them, comparison between vertebrate host proteins and viral
proteins becomes quite interesting. For this comparison, I have
chosen two hemagglutinins of influenza A virus: INF.HEM I and INF.HEM
II [7]. Together, these two hemagglutinins comprise only 550 residues,
and so, there were only three each of Lys-Leu and Leu-Lys. Nevertheless,
it should be noted that within these two hemagglutinins, they were
parts of two pairs of homologous tetrapeptides, as shown in Fig.
1 b. It would also be noted that two of the three Leu-Lys appeared
as Leu-Lys-Ser in INF.HEM II. Thus, the preference of Leu-Lys for
Ser as its carboxyl end partner is truly catholic. The above aroused
interest on the longstanding question of self versus nonself. Confining
ourselves only to Lys-Leu- and Leu-Lys-containing oligopeptides,
how long a fragment of influenza virus hemagglutinins was homologous
with that contained in one or the other of the four vertebrate host
proteins? 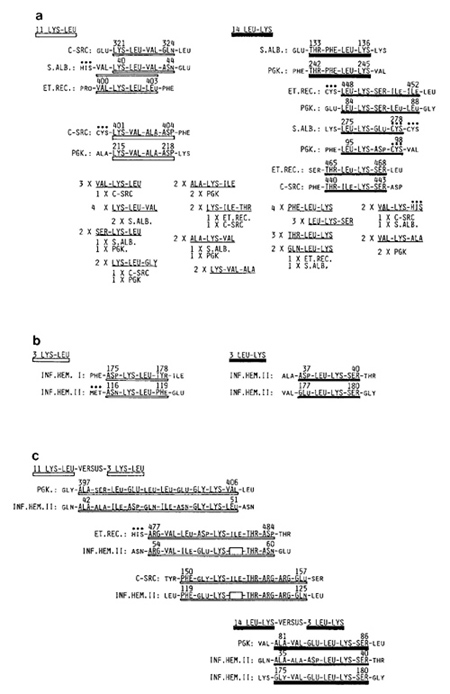
Although there were onla three Lys-Leu in two hemagglutins of influenza a virus, compared to 11 Lys-Leu among the four host proteins, these three Lys-Leu of the virus can also be considered as homologous to six Lys- Val and six Lys-Ile of the host. As shown in Fig. 1 c, the decapeptide ending in Lys-Val of host PGK occupying the 397th-406th positions was seven-tenths homologous with the decapeptide ending in Lys-Leu of INF.HEM II occupying the 42nd-51st positions. In view of the fact that the total number of proteins possessed by the vertebrate host is of the order of 104, it would be no surprise if the decapeptide identical to the above of INF.HEM II were found in at least one unknown host protein. If such is the case, this viral decapeptide is an indisputable self. On the other hand, if the homology of seventenths or thereabouts is the maximal obtainable between this viral peptide fragment and a multitude of host peptide fragments, can it be universally recognized as a nonself? Most instructive concerning this question is the finding reported on human cytotoxic T cell responses to the nuclear matrix protein of influenza A virus [8]. It has been shown that only internal viral proteins, such as the matrix and nucleoproteins of influenza A virus, can invoke a cytotoxic T cell response in infected human and mouse hosts. As far as the matrix protein was concerned, however, it proved incapable of eliciting cytotoxic T cell responses from those human individuals whose class I MHC haplotypes contained HLA-C7 [8]. For those individuals, all peptide fragments of the influenza matrix protein must have appeared as self. Although cytotoxic T cells of HLA-A2 individuals infected with influenza A virus readily responded to the matrix proteins, the test of various peptide fragments revealed that even HLA-A2 cytotoxic T cells recognized only one 19-residue-long peptide fragment representing positions 55- 73 of the matrix protein as nonself [8]. It is probable that positions 42- 51 of INF.HEM II shown in Fig. 1 care the type of peptide fragments that are re cognized as nonself only by helper T cells of particular class 11 MHC haplotypes, thus creating classical responders and nonresponders among individuals. Figure 1 c also shows that two Lys-llecontaining octapeptides of the host ( one derived from ET.REC and the other from C-SRC) enjoyed seven-eighths and sixeighths homology with two heptapeptides of INF.HEM 11, if lIe or Lys-lle of each was deleted. As to Leu-Lys-containing oligopeptides, I shall be content to show only the identical pentapeptide, Val-Glu-Leu-LysSer, shared by PGK of the host and INF.HEM 11. Actually, positions 81-86 are entirely homologous with positions 175-180 of INF.HEM 11. In addition, this PGK hexapeptide was also fivesixths homologous with positions 35~40 of INF.HEM II. ALL Proteins as Divergent Essays Written in One Language During the past several years, we have advanced the notion that all coding sequences in this world are scriptures written in one and the same DNA language [9]. Here, it was shown that the same applies to amino acid sequences of proteins as well. As long as they are written in the same language, two essays on entirely different subjects may have surprisingly many identical and similar components. Witness the following: "The term high ceiling has been used to denote a group of diuretics that have a distinctive action on renal tubular function." "The term high ceiling has been used to denote a group of stocks that show a distinctive pattern of price fluctuations." The first was derived from an essay on diuretic drugs, while the second was from one on stocks and stock markets, yet 15 of the 22 words are identical. Is it a surprise, then, if totally unrelated proteins derived from vertebrates and from a virus share a multitude of identical and homologous oligopeptides? References 1. Bjorkman PJ, Saper MA, Samraouri B, Bennett WS, Strominger JL,
Wiley DC (1987) The foreign antigen binding site and T cell recognition
regions of class I histocompatibility antigens. Nature 329.512-518
|


 ">
"> ">
"> ">
"> ">
"> TESTTESTTESTTESTTESTTESTTESTTESTTESTTES
TTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTEST
TESTTESTTESTTESTTESTTESTTESTTESTTESTTES
TTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTESTTEST