|
1 Physiologisch-Chemisches Institut I der Philipps-Universität, Emil-Mannkopff-Str. 1-2, 3550 Marburg, FRG
The information stored in the DNA of a fertilized egg can be divided
into two different classes: structural information, required for
the synthesis of all macromolecules that build up the organism,
and regulatory information, needed to modulate the expression of
the structural information in time and space, that means during
the development of the different tissues. The connection between
the two types of information is provided by regulatory macromolecules,
that are of course encoded in the structural information and regulate
its expression through interaction with regulatory elements of the
DNA, thus closing the information cycle (Fig. I). The structural
information is stored in the DNA in the form of the genetic code
that was unraveled in the 19605. Part of the structural information
are the signals for initiation and termination of transcription
and translation, as well as the signals for RNA modification and
splicing. On the other hand, little is known about the molecular
mechanisms by which regulatory information is stored in the DNA.
The general idea, however, is that recognition of specific features
of the DNA molecule by regulatory DNA-binding macromolecules is
essential for regulation. What exactly is recognized on the DNA
and how the interaction modulates gene expression are the questions
to be answered. During the past decade, several DNAbinding regulatory
proteins from prokaryotes have been purified to homogeneity, and
their structure as well as their interaction with DNA have been
studied in great detail. A comparison of the amino acid sequence
of 13 DNA-binding regulatory proteins reveals two regions of homology
overlapping the known DNAbinding domains (Fig.2; [ I, 2]). Interestingly,
mutants that disturb the binding of the lac-repressor to the operator
are clustered around these two regions [ 1]. The secondary, tertiary,
and quaternary structure of several DNA-binding regulatory proteins
from bacteria and bacteriophages exhibit striking similarities in
their DNA-binding domains [2]. Not only are these proteins symmetric
dimers or tetramers, but they contain a pair of twofold related
alfa-helices connected by a ß-turn that are responsible for most
of the contacts with the B-form of the DNA double helix. 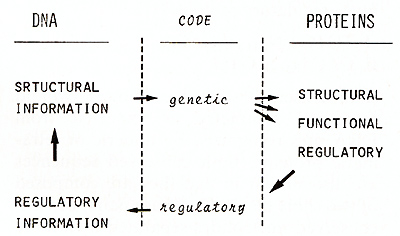 Fig. I. The information cycle  Fig.2. Regions of homology among 13 prokaryotic DNA-binding regulatory proteins One alfa-helix fits into the major groove of DNA while the other
lies across it, holding it in position. If one looks at the relevant
alfa-helices along their longitudinal axis, one observes that the
orientation of the amino acid side chains exhibits a clear polarity.
That means that the nonpolar amino acid side chains are oriented
toward one side of the alfa-helix, whereas the polar and charged
amino acid side chains are oriented toward the other side of the
helix. This would be the site that contacts the DNA major groove.
This brief summary on the structure of prokaryotic regulatory proteins
suggests that a basic protein structure has originated in evolution
that can fulfill the requirements for DNA recognition. The actual
function of a particular regulatory protein may depend on other
domains of the protein that mediate the interaction with different
modulator molecules. As for the DNA sequences that are recognized
by the regulatory proteins, they also show considerable homology.
Two types of conserved sequences can be derived from a comparison
of 23 sites recognized by 13 regulatory proteins [ 1].  Fig. 3 a-c. Structure of the glucocorticoid-binding sites of MMTV and hMTIIA. Computer graphic representation of the DNA double helix containing the nucleotide sequences of a MMTVI; b MMTVIIA ; and c hMTIIA (shown in Fig. 5). The sites of contact with the receptor are indicated by open Iriangles. Those positions hypermethylated in the presence of receptor are marked by full Iriangles. The receptor molecules are represented as broken circles. Numbers refer to the distance from the "cap" site through interaction with intracellular receptors, that in their
turn recognize regulatory eleruents in the neighborhood of the regulated
promoters. Regulatory elements are defined as DNA sequences that
in addition to being required for receptor binding, are needed for
the hormonal regulation of transcription in gene transfer experiments.
They were first reported in the long terminal repeat region (L TR)
of mouse mammary tumor virus (MMTY), that contains the main promoter
for proviral transcription [6-8]. Olucocorticoids were known to
induce viral transcription in different cell lines [9], and gene
transfer experiments with deletion mutants in the L TR region showed
that the sequences relevant for hormonal regulation are located
between 50 and 400 base pairs upstream of the initation of transcription
[7, 10-12]. Within this region, several binding sites for the glucocorticoid
receptor of rat liver have been described [8, 13]. Using a cloned
proviral DNA from OR mice [8], we found four binding sites that
share the hexanucleotide 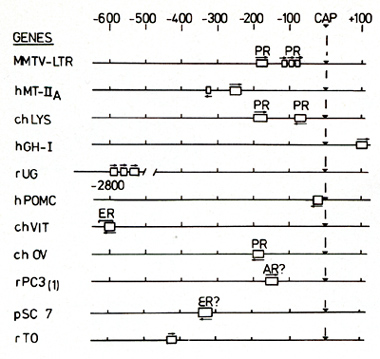
that already mentioned for prokaryotic DNA-binding regulatory
proteins, An analysis of other glucocorticoid-regulated genes showed
that the presence of a regulatory element is not an exclusive property
of the retroviral genome. The human metallothioneine IIA gene (hMTIIA),
that has been shown to be induced by glucocorticoids in many different
cell lines, contains a glucocorticoid regulatory element about 250
base pairs upstream of the initiation of transcription [ 15]. This
element is very similar to the strong binding site found in the
LTR region of MMTV (compare a and c in Fig. 3). In addition, there
is a weak binding site in the hMTIIA promoter located at around
320 base pairs upstream of the initiation of transcription [15].
Similarly to the weak binding site in the L TR region of MMTV (Fig.
3 b), the shorter footprint and methylation protection pattern in
the weak binding site of hMTIIA suggests binding of a receptor monomer.
Interestingly, this weak site at -320 can be deleted without influencing
the hormonal inducibility of hMTIIA [15]. Thus, it could be that
a functional in teraction req uires binding of a receptor dimer
to a strong site on the DNA. In the meantime, we have identified
binding sites for the glucocorticoid receptor in several hormonally
regulated genes. A summary of these results along with data from
the literature is shown in Fig. 4. The promoter for the chicken
lysozyme gene (chL YS), contains two binding sites for the glucocorticoid
receptor, located at around 180 and 60 base pairs upstream of the
initiation of transcription [ 16]. The upper binding site, that
has a lower affinity for the glucocorticoid receptor, coincides
with sequences required for hormone-dependent expression of the
gene in oviduct cells [16]. In fact, these sequences mediate not
only glucocorticoid regulation, but also induction by progesterone
in microinjection experiments [16]. Interestingly, the partially
puri fied progesterone receptor from rabbit uterus binds to the
same sites as the glucocorticoid receptor, although with different
affinity. Thus, it appears that the binding sites for the receptors
of two different steroid hormones may be identical or at least share
common sequences. That these similarities may not be limited to
the progesterone and glucocorticoid receptors is suggested by studies
with genes regulated by other steroid hormones (Fig. 4). The chicken
vitellogenin II gene that is induced by estrogens in the liver,
contains a binding site for the estrogen receptor around 600 nucleotides
upstream of the transcription initiation site [ 17]. An analysis
of the nucleotide sequences in this region reveals an element almost
identical to the binding sites for the glucocorticoid receptor (Fig.
5). A review of the literature showed that a rat gene for a prostatic
protein. that is  Fig.5. Consensus sequence for the glucocorticoid regulatory element. The nucleotide sequences of the main binding sites for the glucocorticoid receptor are aligned to yield maximal homology. Abbreviations are as in Fig. 4 known to be induced by androgens, rPC 3 ( 1 ), also contains a
sequence homologous to the binding site for the glucocorticoid receptor
some 140 nucleotides upstream of the initiation of transcription
([18]; Figs.4 and 5). Finally, an ecdysone-inducible gene of Drosophila
(pSC7) also contains a binding site for the glucocorticoid receptor
some 330 nucleotides upstream of the transcription initiation site
([ 19]; Figs.4 and 5). These findings, taken together, suggest that
the regulatory elements for different steroid hormone receptors
may be similar or at least overlap. The rabbit uteroglobin gene
is induced by glucocorticoids in the lung and by estrogen and progesterone
in the endometrium [20]. We have looked for binding sites for the
glucocorticoid receptor and found none in the neighborhood of the
promoter. The closest binding region detected is located 2700 nucleotides
upstream of the cap site, and is composed of three binding sites
showing sequence homology to other glucocorticoid regulatory elements
(Figs.4 and 5). That this site may be relevant for regulation in
vivo is suggested by the finding of a DNase I hypersensitive site
in this region only in chromatin of hormonally stimulated endometrium
(unpublished results). The human growth hormone gene (hGHI) is induced
by glucocorticoids in several cell lines [21 ]. In fact, gene transfer
experiments with a chimeric gene suggested that a fragment of DNA
containing 500 base pairs upstream of the initiation of transcription
is sufficient for hormonal regulation [22]. In binding experiments
with the glucocorticoid receptor, however, we found amain binding
site located around position + lOO, within the first intron (Figs.4
and 5). If this site is involved in transcriptional regulation in
vivo, it would mean that the regulatory element can act even when
located downstream of the regulated promoter. Taken together, the
data shown in Fig.4 show that the regulatory elements for steroid
hormones share some of the properties of the so-called enhancer
elements [23]. They can act at variable distance from the regulated
promoters, both upstream and downstream, and in both orientations.
There is in fact direct experimental evidence for an enhancer function
of the glucocorticoid regulatory element in the L TR region of MMTV
[7]. A comparison of the nucleotide sequences of ten different binding
sites for the glucocorticoid receptor yields the consensus sequence
shown in Fig. 5. Therefore, the glucocorticoid regulatory elements
have been conserved in evolution between chicken, rodents, and humans.
The bestconserved regions include all those sites that are involved
in direct contacts with the receptor [ 14]. The symmetry in the
element 
base pair has the structure acceptor-donor -acceptor and is therefore symmetric, whereas a GC base pair has the structure acceptor-acceptor-donor (Fig. 6). If one now compares the ten glucocorticoid receptor binding sites with their flanking sequences in terms of this hydrogen bond pattern, one observes a very good preservation of the donor-acceptor structure around the contact sites, with very little agreement outside the binding region (Fig. 6). A certain symmetry can be detected centered at position 10: two well-preserved blocks, 3 to 8 and 12 to 17, separated by less-preserved positions, and interrupted in symmetric positions at 5 and 15. Of course, other interactions are probably implicated in recognition, but the network of hydrogen bonds seems to be an essential part of the code in which regulatory information is stored in DNA. A precise understanding of the molecular mechanisms by which the regulatory code is read could derive from the fine structural analysis of cocrystals containing the DNA-binding domains of regulatory proteins bound to the corresponding nucleotide sequences [27, 28]. Only then will it be possible to decide whether there is a general rule underlying the mechanism of sequence specific recognition by regulatory proteins.
The experimental work reviewed here has been supported by grants
from the Deutsche Forschungsgemeinschaft
1. Gicquel-Sanzey B, Cossart P (1982) EMBO l 1:591-595 2. Takeda Y, Ohlendorf DH, Anderson WF, Matthews BW ( 1983) Science 221: 1020-1026 3. Ogden S, Haggerty D, Stoner CM, Kolodrubetz D, Schleif R (1980) Proc Natl Acad Sci USA77:3346-3350 4. Schmitz A (1981) Nucleic Acid Res 9:277 -291 5. Tjian R (1978) Ce1113: 165-179 6. Geisse S, Scheidereit C, Westphal HM, Hynes NE, aroner B, Beato M (1982) EMBO1I:1613-1619 7. Chandler VL, Maler BA, Yamamoto KR (1983) CeI133:489-499 8. Scheidereit C, Geisse S, Westphal HM, Beato M (1983) Nature 304:749-752 9. Ringold aM (1979) Biochim Biophys Acta 560:487-508 10. Hynes NH, van Ooyen All, Kennedy N, Herrlich P, Ponta H, Groner B (1983) Proc Natl Acad Sci USA 80: 3637-3641 11. Majors l, Varmus HE (1983) Proc Natl Acad Sci USA 80: 5866-5870 12. Buetti E, Diggelmann H (1983) EMBO J 2:1423-1429 13. Payvar F, deFranco DF, Firestone aL, Edgar B, Wrange Ö, Okret S, austafsson lA, Yamamoto KR (1983) CeI135:381-392 14. Scheidereit C, Beato M (1984) Proc Natl Acad Sci USA 81:3029-3033 15. Karin M, Haslinger A, Holtgreve H, Richards RI, Krauter P, Westphal HM, Beato M (1984) Nature 308:513-519 16. Renkawitz R, Schütz a, von der Ahe D, Beato M (1984) CeI137:503-510 17. Jost lP, Seldran M, Geiser M (1984) Proc Natl Acad Sci USA 8:429-433 18. Parker M, Hurst H, Page M (1984) J Steroid Biochem 20:67-71 19. Moritz T, Edström lE, Pongs O (1984) EMBO 13:289-295 20. Beato M, Arnemann J, Menne C, Müller H, Suske a, Wenz M (1983) In: McKerns KW ( ed) Regulation of gene expression by hormones. Plenum, New York, pp 151-175 21. Martial JA, Baxter lD, Goodman HM, Seeburg PH (1977) Proc Natl Acad Sci USA 74: 1816-1820 22. Robins DM, Paek I, Seeburg P, Axel R (1982) Cell29:623-631 23. Banerji J, Rusconi S, Schaffner W (1981) Cell 27: 299- 308 24. Seeman NC, Rosenburg lM, Rich A (1979) Proc Natl Acad Sci USA 72: 804-808 25. Rein R, Kieber-Emmons T, Haydock K, Garduno-Juarez R, Shibata M (1983) J Biomol Struct Dyn 1: 1051-1079 26. Berg oa, Winter RB, von Hippel PH (1981) Biochemistry 20: 6929-6948 27. Anderson l, Ptashne M, Harrison SC (1984) Proc Natl Acad Sci USA 81: 1307-1311 28. Frederick CA, Grable J, Melia M, Samudzi C, Jen-Jacobson L, Wang BC, Greene P, Boyer HW, Rosenberg lM (1984) Nature 309:327-331 |