|
* The experiments summarized in this article were financially supported by grants from the Stiftung Volkswagenwerk and the Deutsche Forschungsgemeinschaft. The Heinrich-Pettelnstitut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium fur Jugend, Familie und Gesundheit, Bonn, FRG A. Introduction To study mechanisms of gene regulation involved in embryonal development, we inserted the Moloney leukemia virus (MMuL V) genome into the germ line of mice. Thirteen different substrains of mice were obtained, each carrying one single copy of the Moloney leukemia virus as a Mendelian gene [ 1-3]. These substrains differ in their genotype ( different chromosomal integration sites; Mov loci) as well as in their phenotype of virus expression: the majority of substrains exhibit no virus expression at all, and four substrains express virus at different stages of development. In Table 1 the characteristics and the time of virus activation during development in the different Mov substrains are summarized. Recent evidence obtained in our laboratory [4] indeed suggested that tissue-specific activation of viral genomes carried in the germ line of mice may be regulated by similar mechanisms, as has been proposed for the tissue-specific activation of developmentally regulated genes [5]. Our results furthermore suggested that the chromosomal position at which virus integration occurred influenced the timing in development and the cell type where the proviral genome became activated [2, 3]. As a means of studying the underlying regulatory mechanisms we have analyzed the extent ofDNA methylation [6-8] of the viral genomes. All proviral genomes carried in the Mov substrains were highly methylated, were not expressed in the tissues tested, and were not infectious in a transfection assay [9]. However, when the methyl groups were removed by molecular cloning of the proviral copies, they were rendered highly infectious [10]. These results strongly suggested that DNA methylation plays a causative role in gene regulation during development and differentiation. The Mov substrains were derived by exposing preimplantation mouse embryos to M -M uL V. Since the infecting retroviral DNA was not methylated, de novo methylation of the proviral genomes must have occurred at some point either during development of the infected embryo and/or as a consequence of their transmission through the germ line. Furthermore, it has been shown previously that early mouse embryos as well as em bryonal carcinoma (EC) cells [11-14], which have many features in common with embryonic ectoderm cells of early mouse embryos, are nonpermissive for replication of M-MuL V. The experiments summarized in this review article were performed to understand the parameters that prevent expression of viral genomes introduced in to early embryos and to correlate this with DNA methylation. Table I. Mouse strains with germ line
integrated Moloney leukemia virus 
Two experimental approaches were used to investigate the molecular parameters that prevent expression of RNA tumor viruses in em bryonal cells. The fate of the infecting viral DNA was directly followed and compared in tissue culture by infecting pluripotent EC cells or differentiated cells. In a second approach the preimplantation or postimplantation mouse embryos were exposed to M-MuL V, and viral genomes car ried in the adult animals derived from the respective infected em bryos were characterized. In both experimental approaches the expression of viral genomes was studied by the XC plaque assay, quantitative RNA hybridization, and/ or in situ hybridization, and modifications of the viral genomes were characterized by restriction enzyme analysis and by transfection assay of the high molecular weight DNA. The results of these experiments have been published [ 15, 16] and will be briefly summarized in Tables 2 and 3. Table 2.De novo methylation of M-MuL V
genomes after infection 
Pluripotent EC cells (F-9 cells) and differentiated cells (EB22/20, a differentiated derivative of EC cells or NIH 3T3 cells) were exposed to M-MuLV [15]. Whereas virus replicated efficiently in the latter cells. as revealed by infectious center assay or RNA hybridization experiments, no virus expression was found in F-9 cells (Table 2). The following experiments were performed to study the block in virus expression in F-9 cells. The kinetics of virus integration were established and indicated that all viral genomes integrated stably into the cellular chromosomal DNA during the first 2 or 3 days after exposure of cells to M-MuLV. Analyses using methylation-sensitive restriction enzymes revealed that viral DNA in F-9 cells remained unmethylated as long as it was in the episomal state but became de novo methylated soon after chromosomal integration. This correlated well with the transfection assay: DNA isolated early after infection was biologically active, whereas DNA isolated late when free viral DNA was no longer present failed to induce XC plaques upon transfection (Table 2). The methylated proviral copies. however, were potentially infectious because they induced XC plaques when the recipient cells for transfection were treated with azacytidine. This drug is believed to interfere with maintenance methylation. In contrast, viral genomes introduced into EB22/20 or NIH 3T3 cells remained unmethylated as well as infectious after chromosomal integration (Table 2). Our results strongly suggest that expression of proviral genomes introduced into pluripotent EC cells is suppressed upon chromosomal integration and that this inactivation can be correlated with de novo methylation of the viral DNA.
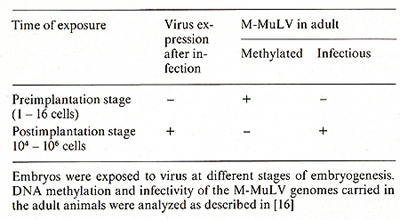
Due to technical problems in obtaining sufficient material, the fate of viral DNA introduced into early mouse embryos could not be analyzed directly in a similar way to that described above for the tissue culture systems. Therefore viral genomes were studied in adult animals derived from the infected embryos. Two stages of embryogenesis, which differ fundamentally in their response to virus infection were compared: (a) Embryos were infected with virus at the preimplantation stage, a stage at which no viral expression takes place [II, 14]: (b) embryos were microinjected with virus at day 8 of gestation. At this stage efficient virus replication occurs in cells of all tissues as revealed by in situ hybridization [16] or by analyzing the tissue distribution of viral DNA and RNA in the adult [17]. The results of analyzing the modification and infectivity of the viral genomes carried in the adults are summarized in Table 3 [16]. Restriction enzyme analysis revealed that copies introduced into preimplantation embryos became de novo methylated and remained highly methylated throughout the life of the animal, whereas viral genomes introduced 5 days later into the postimplantation embryo remained unmethylated. The results of transfection assays con firmed these results. DNA derived from animals exposed to virus at the postimplantation stage was highly infectious, in contrast to DNA from animals exposed to virus at the preimplantation stage. These observations extend the results obtained in vitro with EC cells to the in vivo situation. They suggest that an efficient de novo methylation activity is a characteristic of totipotent early embryos and may be involved in the inhibition of viral gene expression. Neither de novo methylation activity nor inhibition of virus replication, however, is observed at day 8 of development.
The introduction of foreign cellular and retroviral genomes into early mouse embryos has been used as a means of investigating the regulation of gene expression in mammalian development [ 18-23]. The results obtained in our system established that both embryonal carcinoma cells and preimplantation mouse embryos are nonpermissive for expression of retroviral genomes. Retroviruses introduced into differentiated derivatives of EC cells or into postimplantation mouse embryos at day 8 of gestation, however, were able to replicate efficiently. This defines a switch ofearly differentiating cells in their ability to support retroviral expression which is developmentally regulated. The switch in gene expression was correlated with efficient de novo methylase activity in pluripotent cells. Retroviral genomes introduced into EC cells or into preimplantation mouse embryos became efficiently de novo methylated, in contrast to viral genomes introduced into differentiated cells or into postimplantation embryos. The results with EC cells indicated that this enzyme activity de novo methylates viral genomes only after chromosomal integration and does not act on DNA molecules which are in the episomal state. This is relevant to the observation that DNA microinjected into mouse zygotes [24] or into Xenopu.s eggs [25] is expressed as long as it remains in an episomal state. In addition, unintegrated DNA injected into Xenopus eggs was shown to remain unmethylated [26]. Our results furthermore show that the maintenance methylation activity is faithful in preserving the respective methylation pattern of the proviral genomes throughout the life of the animal. The de novo methylation activity in embryonal cells may be of general significance as not only viral but also cloned globin DNA, which was microinjected into mouse zygotes, became de novo methylated (F. Costantini and E. Lacy, personal comminication). If the de novo methylation activity in embryonal and efficient maintenance methylation in later cells are involved in repression of proviral genomes, what is the origin of infectious virus in mice derived from preimplantation embryos exposed to virus? Because virus, once activated. will infect all susceptible cells and spread throughout the animal, demethylation and activation of virus at a given stage of development and in a specific. as yet unidentified, population of cells would be sufficient to lead to viremia. Demethylation of a given provirus in specific cells may depend on the chromosomal position where integration took place, and proviral genome activation may thus be regulated by similar mechanisms as has been proposed for the tissue-specific activation of developmentally regulated genes [5, 27]. Gene activation of the proviral genome in Mov-l mice appears to be compatible with such a hypothesis [4]. Our results suggest that embryonal cells may possess an efficient de novo methylation activity that inactivates any DNA which is introduced into the early embryo. This may have evolved as a mechanism to protect the developing embryo against deleterious conseq uences of virus infections. Finally. our results pose intriguing questions concerning the control of gene expression during early development, and it will be of great interest to study the methylation of genes that are active in preimplantation embryos and in embryonal carcinoma cells.
I. Jaenisch R (1976) Proc Natl Acad Sci USA 73: 1260 |